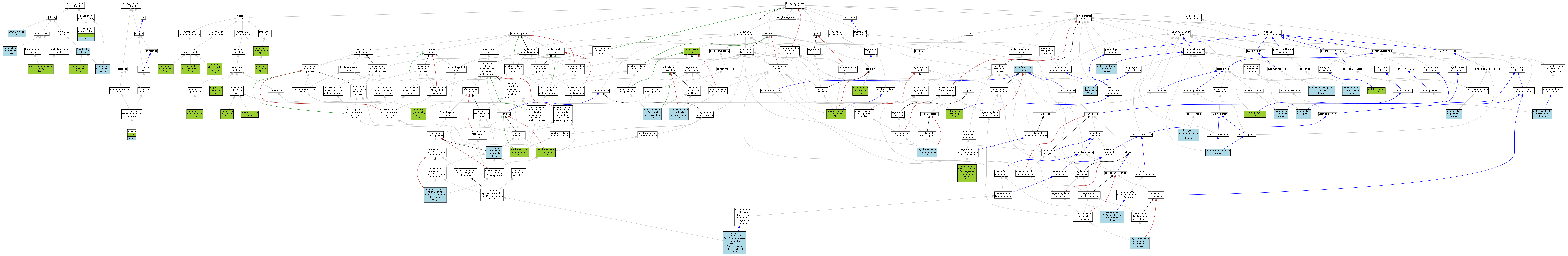
| Category | ID | Term | Human | Mouse | Rat | Chicken | Zfish | Fly | Worm | Dicty | Dicot | Yeast | Pombe | Ecoli |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0005488 | binding | X | Mouse | X | X | X | X | X | X | Dicot | X | X | X |
| Molecular Function | GO:0030528 | transcription regulator activity | X | Mouse | X | X | X | X | X | X | Dicot | X | X | X |
| Cellular Component | GO:0005623 | cell | X | Mouse | X | X | X | X | X | X | Dicot | X | X | X |
| Cellular Component | GO:0044464 | cell part | X | Mouse | X | X | X | X | X | X | Dicot | X | X | X |
| Cellular Component | GO:0043226 | organelle | X | Mouse | X | X | X | X | X | X | Dicot | X | X | X |
| Biological Process | GO:0065007 | biological regulation | X | Mouse | X | X | X | X | X | X | Dicot | X | X | X |
| Biological Process | GO:0009987 | cellular process | X | Mouse | X | X | X | X | X | X | Dicot | X | X | X |
| Biological Process | GO:0032502 | developmental process | X | Mouse | X | X | X | X | X | X | Dicot | X | X | X |
| Biological Process | GO:0040007 | growth | X | Mouse | X | X | X | X | X | X | Dicot | X | X | X |
| Biological Process | GO:0008152 | metabolic process | X | Mouse | X | X | X | X | X | X | Dicot | X | X | X |
| Biological Process | GO:0032501 | multicellular organismal process | X | Mouse | X | X | X | X | X | X | Dicot | X | X | X |
| Biological Process | GO:0048519 | negative regulation of biological process | X | Mouse | X | X | X | X | X | X | Dicot | X | X | X |
| Biological Process | GO:0048518 | positive regulation of biological process | X | Mouse | X | X | X | X | X | X | Dicot | X | X | X |
| Biological Process | GO:0050789 | regulation of biological process | X | Mouse | X | X | X | X | X | X | Dicot | X | X | X |
| Biological Process | GO:0000003 | reproduction | X | Mouse | X | X | X | X | X | X | Dicot | X | X | X |
| Biological Process | GO:0022414 | reproductive process | X | Mouse | X | X | X | X | X | X | Dicot | X | X | X |
| Biological Process | GO:0050896 | response to stimulus | X | Mouse | X | X | X | X | X | X | Dicot | X | X | X |
| Category | ID | Term | DB Object | Evidence | With_From | Organism | Reference |
|---|---|---|---|---|---|---|---|
| Molecular Function | GO:0003682 | chromatin binding | Dlx1 | IDA | Mouse | MGI:MGI:3032902|PMID:14769946 | |
| Molecular Function | GO:0003677 | DNA binding | Dlx1 | IDA | Mouse | MGI:MGI:3032902|PMID:14769946 | |
| Molecular Function | GO:0003677 | DNA binding | Dlx1 | IDA | Mouse | MGI:MGI:1100181|PMID:9415433 | |
| Molecular Function | GO:0003677 | DNA binding | Nkx3-1 | IDA | Mouse | MGI:MGI:1928182|PMID:10993896 | |
| Molecular Function | GO:0042803 | protein homodimerization activity | AT5G65310 | IDA | Dicot | TAIR:Publication:501680370|PMID:11247607 | |
| Molecular Function | GO:0042803 | protein homodimerization activity | AT4G16780 | IDA | Dicot | TAIR:Publication:5116|PMID:8253077 | |
| Molecular Function | GO:0043565 | sequence-specific DNA binding | AT4G40060 | IDA | Dicot | TAIR:Publication:501680370|PMID:11247607 | |
| Molecular Function | GO:0043565 | sequence-specific DNA binding | AT5G15150 | IDA | Dicot | TAIR:Publication:501680370|PMID:11247607 | |
| Molecular Function | GO:0043565 | sequence-specific DNA binding | AT5G65310 | IDA | Dicot | TAIR:Publication:501680370|PMID:11247607 | |
| Molecular Function | GO:0043565 | sequence-specific DNA binding | AT4G16780 | IDA | Dicot | TAIR:Publication:5116|PMID:8253077 | |
| Molecular Function | GO:0016563 | transcription activator activity | Nkx3-1 | IDA | Mouse | MGI:MGI:1928182|PMID:10993896 | |
| Molecular Function | GO:0016563 | transcription activator activity | AT3G61890 | IDA | Dicot | TAIR:Publication:501717432|PMID:16055682 | |
| Molecular Function | GO:0016563 | transcription activator activity | AT4G40060 | IDA | Dicot | TAIR:Publication:501717432|PMID:16055682 | |
| Molecular Function | GO:0016563 | transcription activator activity | AT5G65310 | IDA | Dicot | TAIR:Publication:501717432|PMID:16055682 | |
| Molecular Function | GO:0003700 | transcription factor activity | Nkx3-1 | IDA | Mouse | MGI:MGI:1928182|PMID:10993896 | |
| Molecular Function | GO:0008134 | transcription factor binding | Nkx3-1 | IDA | Mouse | MGI:MGI:1928182|PMID:10993896 | |
| Cellular Component | GO:0005634 | nucleus | Dlx1 | IDA | Mouse | MGI:MGI:2389286|PMID:12397111 | |
| Cellular Component | GO:0005634 | nucleus | Dlx1 | IDA | Mouse | MGI:MGI:2662644|PMID:12724837 | |
| Cellular Component | GO:0005634 | nucleus | Dlx1 | IDA | Mouse | MGI:MGI:1346965|PMID:10516593 | |
| Cellular Component | GO:0005634 | nucleus | Nkx3-1 | IDA | Mouse | MGI:MGI:3603077|PMID:15987773 | |
| Cellular Component | GO:0005634 | nucleus | AT3G61890 | IDA | Dicot | TAIR:Publication:501713186|PMID:15369784 | |
| Cellular Component | GO:0005634 | nucleus | AT4G16780 | IDA | Dicot | TAIR:Publication:501714452|PMID:15610358 | |
| Biological Process | GO:0048646 | anatomical structure formation | Dlx6 | IGI | MGI:MGI:101926 | Mouse | MGI:MGI:3765253|PMID:17878916 |
| Biological Process | GO:0048754 | branching morphogenesis of a tube | Nkx3-1 | IMP | MGI:MGI:1928655 | Mouse | MGI:MGI:1891543|PMID:11002344 |
| Biological Process | GO:0030154 | cell differentiation | Dlx1 | IMP | MGI:MGI:1857826 | Mouse | MGI:MGI:2389286|PMID:12397111 |
| Biological Process | GO:0008283 | cell proliferation | AT4G16780 | IMP | Dicot | TAIR:Publication:1211|PMID:10477292 | |
| Biological Process | GO:0021893 | cerebral cortex GABAergic interneuron fate commitment | Dlx1 | IGI | MGI:MGI:94902 | Mouse | MGI:MGI:3775335|PMID:17678855 |
| Biological Process | GO:0030326 | embryonic limb morphogenesis | Dlx6 | IGI | MGI:MGI:101926 | Mouse | MGI:MGI:2181241|PMID:12112878 |
| Biological Process | GO:0048706 | embryonic skeletal development | Dlx1 | IGI | MGI:MGI:94902 | Mouse | MGI:MGI:87771|PMID:9187081 |
| Biological Process | GO:0048706 | embryonic skeletal development | Dlx1 | IMP | MGI:MGI:1857826 | Mouse | MGI:MGI:87771|PMID:9187081 |
| Biological Process | GO:0030855 | epithelial cell differentiation | Dlx6 | IGI | MGI:MGI:101926 | Mouse | MGI:MGI:3765253|PMID:17878916 |
| Biological Process | GO:0042472 | inner ear morphogenesis | Dlx6 | IGI | MGI:MGI:101926 | Mouse | MGI:MGI:3689133|PMID:16900517 |
| Biological Process | GO:0030308 | negative regulation of cell growth | AT4G40060 | IMP | Dicot | TAIR:Publication:501711407|PMID:14623244 | |
| Biological Process | GO:0050680 | negative regulation of epithelial cell proliferation | Nkx3-1 | IMP | MGI:MGI:1928655 | Mouse | MGI:MGI:1891543|PMID:11002344 |
| Biological Process | GO:0043524 | negative regulation of neuron apoptosis | Dlx1 | IMP | MGI:MGI:1857826 | Mouse | MGI:MGI:3604035|PMID:16007083 |
| Biological Process | GO:0048715 | negative regulation of oligodendrocyte differentiation | Dlx1 | IGI | MGI:MGI:94902 | Mouse | MGI:MGI:3775335|PMID:17678855 |
| Biological Process | GO:0016481 | negative regulation of transcription | AT4G16780 | IDA | Dicot | TAIR:Publication:1211|PMID:10477292 | |
| Biological Process | GO:0000122 | negative regulation of transcription from RNA polymerase II promoter | Dlx1 | IGI | MGI:MGI:94902 | Mouse | MGI:MGI:3775335|PMID:17678855 |
| Biological Process | GO:0042475 | odontogenesis of dentine-containing tooth | Dlx1 | IGI | MGI:MGI:94902 | Mouse | MGI:MGI:87771|PMID:9187081 |
| Biological Process | GO:0048573 | photoperiodism, flowering | AT4G40060 | IMP | Dicot | TAIR:Publication:501711407|PMID:14623244 | |
| Biological Process | GO:0050679 | positive regulation of epithelial cell proliferation | Dlx6 | IGI | MGI:MGI:101926 | Mouse | MGI:MGI:3689133|PMID:16900517 |
| Biological Process | GO:0045941 | positive regulation of transcription | AT3G61890 | IDA | Dicot | TAIR:Publication:501717432|PMID:16055682 | |
| Biological Process | GO:0045941 | positive regulation of transcription | AT4G40060 | IDA | Dicot | TAIR:Publication:501717432|PMID:16055682 | |
| Biological Process | GO:0045941 | positive regulation of transcription | AT5G65310 | IDA | Dicot | TAIR:Publication:501717432|PMID:16055682 | |
| Biological Process | GO:0030850 | prostate gland development | Nkx3-1 | IMP | MGI:MGI:1928654 | Mouse | MGI:MGI:1861552|PMID:10906459 |
| Biological Process | GO:0030850 | prostate gland development | Nkx3-1 | IMP | MGI:MGI:1926960 | Mouse | MGI:MGI:1335939|PMID:10215624 |
| Biological Process | GO:0030850 | prostate gland development | Nkx3-1 | IMP | MGI:MGI:1928655 | Mouse | MGI:MGI:1891543|PMID:11002344 |
| Biological Process | GO:0009954 | proximal/distal pattern formation | Dlx1 | IMP | MGI:MGI:1857826 | Mouse | MGI:MGI:87771|PMID:9187081 |
| Biological Process | GO:0009954 | proximal/distal pattern formation | Dlx1 | IGI | MGI:MGI:94902 | Mouse | MGI:MGI:87771|PMID:9187081 |
| Biological Process | GO:0010017 | red or far red light signaling pathway | AT4G16780 | IEP | Dicot | TAIR:Publication:501723260|PMID:17965713 | |
| Biological Process | GO:0048510 | regulation of timing of transition from vegetative to reproductive phase | AT4G40060 | IMP | Dicot | TAIR:Publication:501711407|PMID:14623244 | |
| Biological Process | GO:0021882 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment | Dlx1 | IGI | MGI:MGI:94902 | Mouse | MGI:MGI:3775335|PMID:17678855 |
| Biological Process | GO:0006355 | regulation of transcription, DNA-dependent | Nkx3-1 | IDA | Mouse | MGI:MGI:1928182|PMID:10993896 | |
| Biological Process | GO:0009737 | response to abscisic acid stimulus | AT5G65310 | IDA | Dicot | TAIR:Publication:501705889|PMID:12678559 | |
| Biological Process | GO:0009737 | response to abscisic acid stimulus | AT5G65310 | IMP | Dicot | TAIR:Publication:501705889|PMID:12678559 | |
| Biological Process | GO:0009737 | response to abscisic acid stimulus | AT3G61890 | IMP | Dicot | TAIR:Publication:501725252|PMID:15604708 | |
| Biological Process | GO:0009737 | response to abscisic acid stimulus | AT3G61890 | IEP | Dicot | TAIR:Publication:2556|PMID:9617808 | |
| Biological Process | GO:0009646 | response to absence of light | AT5G53980 | IEP | Dicot | TAIR:Publication:501717432|PMID:16055682 | |
| Biological Process | GO:0009733 | response to auxin stimulus | AT4G16780 | IMP | Dicot | TAIR:Publication:1211|PMID:10477292 | |
| Biological Process | GO:0009733 | response to auxin stimulus | AT3G01220 | IEP | Dicot | TAIR:Publication:501683539|PMID:12644682 | |
| Biological Process | GO:0009733 | response to auxin stimulus | AT4G36740 | IEP | Dicot | TAIR:Publication:501714446|PMID:15567172 | |
| Biological Process | GO:0009637 | response to blue light | AT4G40060 | IMP | Dicot | TAIR:Publication:501711407|PMID:14623244 | |
| Biological Process | GO:0009637 | response to blue light | AT5G53980 | IEP | Dicot | TAIR:Publication:501717432|PMID:16055682 | |
| Biological Process | GO:0009735 | response to cytokinin stimulus | AT4G16780 | IEP | Dicot | TAIR:Publication:501717764|PMID:16212609 | |
| Biological Process | GO:0010218 | response to far red light | AT4G16780 | IEP | Dicot | TAIR:Publication:5118|PMID:8106086 | |
| Biological Process | GO:0006970 | response to osmotic stress | AT3G61890 | IEP | Dicot | TAIR:Publication:2556|PMID:9617808 | |
| Biological Process | GO:0009651 | response to salt stress | AT3G61890 | IGI | Dicot | TAIR:Publication:501713186|PMID:15369784 | |
| Biological Process | GO:0048364 | root development | AT4G16780 | IMP | Dicot | TAIR:Publication:1211|PMID:10477292 | |
| Biological Process | GO:0007431 | salivary gland development | Nkx3-1 | IMP | MGI:MGI:1928654 | Mouse | MGI:MGI:1861552|PMID:10906459 |
| Biological Process | GO:0009641 | shade avoidance | AT4G16780 | IEP | Dicot | TAIR:Publication:501718653|PMID:16565297 | |
| Biological Process | GO:0010016 | shoot morphogenesis | AT4G16780 | IMP | Dicot | TAIR:Publication:5233|PMID:8449400 | |
| Biological Process | GO:0009826 | unidimensional cell growth | AT4G16780 | IMP | Dicot | TAIR:Publication:1211|PMID:10477292 |
| IC Inferred by curator |
| IDA Inferred from direct assay |
| IEA Inferred from electronic annotation |
| IEP Inferred from expression pattern |
| IGC Inferred from genomic context |
| IGI Inferred from genetic interaction |
| IMP Inferred from mutant phenotype |
| IPI Inferred from physical interaction |
| ISA Inferred from sequence alignment |
| ISM Inferred from sequence model |
| ISO Inferred from sequence orthology |
| ISS Inferred from sequence or structural similarity |
| NAS Non-traceable Author Statement |
| ND No biological data available |
| RCA Reviewed computational analysis |
| TAS Traceable author statement |
| NR Not recorded (appears in some legacy annotations) |