Phenotypes associated with this allele
|
|
| Find Mice |
Using the International Mouse Strain Resource (IMSR)
Mouse lines carrying:
Vangl1Gt(XL802)Byg mutation
(1 available);
any
Vangl1 mutation
(85 available)
|
|
|

Abnormal orientation of hair cells of the cochlea in Vangl1Gt(XL802)Byg/Vangl1Gt(XL802)Byg embryos
hearing/vestibular/ear
|
|
• modest but significant misorientation in all hair cell layers
• however, the number and organization of hair cell rows is not different from wild-type controls
|
cardiovascular system
|
N |
• unlike mice homozygous for Vangl2Lp alone, nor outflow tract abnormalities are detected
|
nervous system
|
N |
• unlike mice with mutations in Vangl2, no neural tube defects are detected
|
|
|
• modest but significant misorientation in all hair cell layers
• however, the number and organization of hair cell rows is not different from wild-type controls
|
|
|
| Find Mice |
Using the International Mouse Strain Resource (IMSR)
Mouse lines carrying:
Vangl1Gt(XL802)Byg mutation
(1 available);
any
Vangl1 mutation
(85 available)
Vangl2ska17 mutation
(0 available);
any
Vangl2 mutation
(34 available)
|
|
|
normal phenotype
|
|
• no tail, neural tube or eyelid fusion defects are detected
|
reproductive system
nervous system
|
|
• inner hair cells are misoriented compared to in wild-type mice
|
|
|
• outer hair cells are misoriented compared to in wild-type mice
|
limbs/digits/tail
|
|
• compared with Vangl1Gt(XL802)Byg/Vangl1Gt(XL802)Byg Vangl2tm1.2Yy/Vangl2+
|
hearing/vestibular/ear
|
|
• inner hair cells are misoriented compared to in wild-type mice
|
|
|
• outer hair cells are misoriented compared to in wild-type mice
|
|
|
| Find Mice |
Using the International Mouse Strain Resource (IMSR)
Mouse lines carrying:
Vangl1Gt(XL802)Byg mutation
(1 available);
any
Vangl1 mutation
(85 available)
Vangl2tm1.2Yy mutation
(0 available);
any
Vangl2 mutation
(34 available)
|
|
|
embryo
|
|
• positioning of the posterior notochord (node) cilia is randomized unlike in wild-type mice
• in the posterior notochord (node), cell density is slightly reduced and cilia localization is altered compared to in wild-type mice
|
|
|
• nodal flow produces swirling vortices and time to cross the posterior notochord (node) is lengthened compared to in wild-type mice
• unilateral nodal flow in the posterior notochord (node) is compromised compared to in wild-type mice
• however, cilia in the posterior notochord beat at the same speed and direction as in wild-type mice
|
|
|
• embryos fail to turn unlike wild-type mice
|
|
|
• cell density and the number of posteriorly localized basal bodies in the posterior notochord (node) are decreased compared to in wild-type mice
|
growth/size/body
|
|
• embryos exhibit a reduction in length to width ratio compared with wild-type mice
|
|
|
• mice exhibit left-right asymmetry defects compared with wild-type mice
• however, nodal cilia appear normal
|
nervous system
|
|
• inner hair cells are misoriented compared to in wild-type mice
|
|
|
• outer hair cells are misoriented compared to in wild-type mice
|
cardiovascular system
respiratory system
hearing/vestibular/ear
|
|
• inner hair cells are misoriented compared to in wild-type mice
|
|
|
• outer hair cells are misoriented compared to in wild-type mice
|
cellular
|
|
• positioning of the posterior notochord (node) cilia is randomized unlike in wild-type mice
• in the posterior notochord (node), cell density is slightly reduced and cilia localization is altered compared to in wild-type mice
|
|
|
• nodal flow produces swirling vortices and time to cross the posterior notochord (node) is lengthened compared to in wild-type mice
• unilateral nodal flow in the posterior notochord (node) is compromised compared to in wild-type mice
• however, cilia in the posterior notochord beat at the same speed and direction as in wild-type mice
|
|
|
| Find Mice |
Using the International Mouse Strain Resource (IMSR)
Mouse lines carrying:
Vangl1Gt(XL802)Byg mutation
(1 available);
any
Vangl1 mutation
(85 available)
Vangl2tm1.2Yy mutation
(0 available);
any
Vangl2 mutation
(34 available)
|
|
|
nervous system
|
|
• outer hair cells in rows 3 and 2 are misoriented compared to in wild-type mice
|
limbs/digits/tail
|
|
• compared with Vangl2Lp/Vangl2Lp Vangl2tm1.2Yy/Vangl2+
|
hearing/vestibular/ear
|
|
• outer hair cells in rows 3 and 2 are misoriented compared to in wild-type mice
|
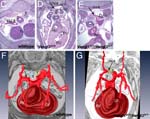
Vangl1Gt(XL802)Byg/Vangl1+ Vangl2Lp/Vangl2+ and Vangl2Lp/Vangl2Lp embryos exhibit aberrant right subclavian artery
mortality/aging
|
|
• fewer than expected double heterozygotes are found at weaning (20% rather than the expected 50%) given the presence of craniorachischisis late embryonic lethality is probably the cause of the distorted ratio
|
nervous system
|
|
• seen in over 60% of double heterozygotes at E13.5 - E18.5
• phenotype is as severe as in mice homozygous for Vangl2Lp alone
• no obvious neural tube defects are seen in surviving mice
|
|
|
• 20% of inner hair cell bundles are misoriented
|
|
|
• some bundles in all 3 layers are misoriented especially at the apical turn (over 50% of bundles in OHC1, 65% in OHC2, over 80% in OHC3)
• vertices are randomly oriented with rotation angles of 40 - 180 degrees
|
cardiovascular system
|
N |
• unlike mice homozygous for Vangl2Lp alone, no outflow tract abnormalities are detected in double heterozygotes
|
|
|
• at E14.5, the right subclavian artery is positioned dorsal to the esophagus
|
hearing/vestibular/ear
|
|
• reduced in size at E18.5 in mice displaying craniorachischisis
|
|
|
• 20% of inner hair cell bundles are misoriented
|
|
|
• some bundles in all 3 layers are misoriented especially at the apical turn (over 50% of bundles in OHC1, 65% in OHC2, over 80% in OHC3)
• vertices are randomly oriented with rotation angles of 40 - 180 degrees
|
limbs/digits/tail
embryo
|
|
• seen in over 60% of double heterozygotes at E13.5 - E18.5
• phenotype is as severe as in mice homozygous for Vangl2Lp alone
• no obvious neural tube defects are seen in surviving mice
|



 Analysis Tools
Analysis Tools
