Phenotypes associated with this allele
Allelic
Composition |
Sptbn1tm1Mish/Sptbn1tm1Mish
|
|
Genetic
Background |
either: (involves: 129S6/SvEvTac * C57BL/6) or (involves: 129S6/SvEvTac * NIH Black Swiss) |
|
| Find Mice |
Using the International Mouse Strain Resource (IMSR)
Mouse lines carrying:
Sptbn1tm1Mish mutation
(0 available);
any
Sptbn1 mutation
(139 available)
|
|
|
mortality/aging
|
|
• abnormal or degenerating embryos detected between E8.5 and E16.5
|
cardiovascular system
|
|
• lack of branching network of vessels in yolk sac
|
|
|
• absence of trabeculated pattern of myocardial tissue; myoblasts were hyperplastic and lacked linear arrangement of nuclei
|
|
|
• small ventricular lumen; occlusion of atrioventricular region
|
craniofacial
digestive/alimentary system
embryo
|
|
• lack of branching network of vessels in yolk sac
|
|
|
• smaller body and head size, evident at E9.5
|
growth/size/body
|
|
• smaller body and head size, evident at E9.5
|
liver/biliary system
muscle
|
|
• absence of trabeculated pattern of myocardial tissue; myoblasts were hyperplastic and lacked linear arrangement of nuclei
|
nervous system
|
|
• abnormal anatomy of primary brain vesicles
|
Allelic
Composition |
Sptbn1tm1Mish/Sptbn1+
|
|
Genetic
Background |
involves: 129S6/SvEvTac |
|
| Find Mice |
Using the International Mouse Strain Resource (IMSR)
Mouse lines carrying:
Sptbn1tm1Mish mutation
(0 available);
any
Sptbn1 mutation
(139 available)
|
|
|
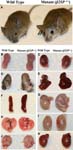
Beckwith-Wiedemann Syndrome-like phenotypes in Sptbn1tm1Mish/Spnb2+ mice
mortality/aging
|
|
• increase in incidence of sudden death in males
|
growth/size/body
|
|
• 25% increase in average body size and mass compared to wild-type
|
neoplasm
|
|
• adrenocortical carcinomas
|
 |
• 3.4% incidence of epithelial tumors in the ovary
|
 |
• 3.4% incidence of testicular carcinomas
|
|
|
• 37.9-40% increase in incidence of liver tumors
|
|
|
• 3.4% incidence of breast adenomas
|
|
|
• 6.9% incidence of spleen lymphomas
|
|
|
• pancreatic carcinomas and adrenocortical carcinomas
• 3.4% incidence of testicular carcinomas
|
|
|
• 6.9% incidence of adenocarcinomas in the lung
|
|
|
• 10.3% incidence of renal cell carcinomas
|
|
|
• 3.4% incidence of abdominal/mesenchymal sarcomas
|
hearing/vestibular/ear
integument
cardiovascular system
craniofacial
digestive/alimentary system
endocrine/exocrine glands
|
|
• adrenocortical carcinomas
|
 |
• 3.4% incidence of epithelial tumors in the ovary
|
 |
• 3.4% incidence of testicular carcinomas
|
liver/biliary system
|
|
• 37.9-40% increase in incidence of liver tumors
|
renal/urinary system
|
|
• 10.3% incidence of renal cell carcinomas
|
reproductive system
 |
• 3.4% incidence of epithelial tumors in the ovary
|
 |
• 3.4% incidence of testicular carcinomas
|
respiratory system
|
|
• 6.9% incidence of adenocarcinomas in the lung
|
nervous system
Allelic
Composition |
Sptbn1tm1Mish/Sptbn1+
|
|
Genetic
Background |
involves: 129S6/SvEvTac * C57BL/6 |
|
| Find Mice |
Using the International Mouse Strain Resource (IMSR)
Mouse lines carrying:
Sptbn1tm1Mish mutation
(0 available);
any
Sptbn1 mutation
(139 available)
|
|
|
neoplasm
|
|
• mice spontaneously develop tumors of the liver with an incidence of 40%
• lesions included early centrilobular steatosis and dysplasia with nuclear disarray and stratification
• many of these lesions progress to poorly differentiated carcinomas
|
liver/biliary system
|
|
• apoptosis rates are lower among hepatocytes potentially leading to lesion formation
|
|
|
• mice spontaneously develop tumors of the liver with an incidence of 40%
• lesions included early centrilobular steatosis and dysplasia with nuclear disarray and stratification
• many of these lesions progress to poorly differentiated carcinomas
|
cellular
|
|
• apoptosis rates are lower among hepatocytes potentially leading to lesion formation
|
|
|
| Find Mice |
Using the International Mouse Strain Resource (IMSR)
Mouse lines carrying:
Itih4tm1Mish mutation
(0 available);
any
Itih4 mutation
(60 available)
Sptbn1tm1Mish mutation
(0 available);
any
Sptbn1 mutation
(139 available)
|
|
|
neoplasm
|
|
• only 1 mouse in 25 developed a liver neoplasm compared to the 40% incidence rate observed in mice carrying just one null allele for Spnb2
• the lone tumor was about 10-fold smaller than the mean tumor sizes observed in the control mice
|
liver/biliary system
|
|
• apoptosis rates are lower in hepatocytes compared to control mice
|
|
|
• hepatocyte proliferation is lower than in controls
|
|
|
• only 1 mouse in 25 developed a liver neoplasm compared to the 40% incidence rate observed in mice carrying just one null allele for Spnb2
• the lone tumor was about 10-fold smaller than the mean tumor sizes observed in the control mice
|
cellular
|
|
• apoptosis rates are lower in hepatocytes compared to control mice
|
|
|
• hepatocyte proliferation is lower than in controls
|



 Analysis Tools
Analysis Tools

