Phenotypes associated with this allele
|
|
| Find Mice |
Using the International Mouse Strain Resource (IMSR)
Mouse lines carrying:
Pth2tm1Vlcg mutation
(0 available);
any
Pth2 mutation
(4 available)
Tg(CAG-LacZ,-Pth2)1Tbu mutation
(0 available)
Tg(Prm-cre)58Og mutation
(4 available)
|
|
|
reproductive system
|
N |
• male fertility is restored
|
normal phenotype
 |
• mice are viable and fertile with no gross abnormalities and expression levels of Ogt that are similar to controls suggesting inactivation of the paternal X chromosome in all tissues and cell types surveyed
• mice do not produce any offspring carrying the recombined allele
|
|
|
| Find Mice |
Using the International Mouse Strain Resource (IMSR)
Mouse lines carrying:
Casktm1.1Sud mutation
(0 available);
any
Cask mutation
(14 available)
Casktm1Sud mutation
(1 available);
any
Cask mutation
(14 available)
Tg(Prm-cre)58Og mutation
(4 available)
|
|
|
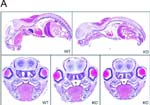
Cleft palate in Casktm1Sud/Casktm1.1Sud Tg(Prm-cre)580g/0 mice
mortality/aging
 |
• die within a few hours of birth
|
craniofacial
 |
• observed in homozygotes with ~80% penetrance
|
digestive/alimentary system
 |
• observed in homozygotes with ~80% penetrance
|
nervous system
 N N |
• synapse formation and membrane properties of cultured neurons are not significantly different from wild-type neurons; vesicle pool sizes and evoked neurotransmitter release are similar in mutant and wild-type neurons
|
 |
• number of apoptotic neurons in thalamus is increased 3-fold compared to controls
|
 |
• excitatory spontaneous mini events show 2-fold increase in frequency in cultured neurons, while inhibitory minifrequency is decreased ~1.4 fold
|
respiratory system
 |
• plethysmography of newborns shows severe postnatal respiratory failure
|
cellular
 |
• number of apoptotic neurons in thalamus is increased 3-fold compared to controls
|
growth/size/body
 |
• observed in homozygotes with ~80% penetrance
|
|
|
| Find Mice |
Using the International Mouse Strain Resource (IMSR)
Mouse lines carrying:
Meig1tm1.1Zzha mutation
(0 available);
any
Meig1 mutation
(14 available)
Tg(Prm-cre)58Og mutation
(4 available)
|
|
|
reproductive system
 N N |
• all males are fertile and sire normal numbers of offspring
• testis weight to body weight and seminal vesicle weight to body weight ratios are normal
• epididymidal sperm count, testis as well as epididymal histology and sperm morphology are normal, and no reduction in sperm motility is observed
|
|
|
| Find Mice |
Using the International Mouse Strain Resource (IMSR)
Mouse lines carrying:
Fyco1tm1.1Arte mutation
(0 available);
any
Fyco1 mutation
(44 available)
Tg(Prm-cre)58Og mutation
(4 available)
|
|
|
vision/eye
|
|
• at P60, lens epithelial cells show extensive vacuolization
|
|
|
• at P60, lens fiber cells are large and display irregular shapes
• at P0, P7, and P14, differentiating lens fiber cells show abnormal retention of cellular organelles, including endoplasmic reticulum (ER), mitochondria, and Golgi apparatus (GA), suggesting impaired organelle removal
|
|
|
• mice show initial signs of lens opacities at 4 weeks that develop into mild cataracts at ~8 weeks of age
|
|
|
• mice develop mature bilateral cataracts by 16 weeks of age; extent of lens opacities is variable among mice
|
cellular
|
|
• at P14, lenses show an increased Golgi apparatus mass relative to age-matched wild-type lenses
|
|
|
• at P0, P7, P14 and P21, lenses show an increased ER mass relative to age-matched wild-type lenses
|
|
|
• at P0, lenses show alterations in many autophagy-associated genes, proteins and lipids, including a 1.48-fold increase in the level of SQSTM1/p62 and a modest accumulation of phosphatidylethanolamine (PE) phospholipids, suggesting impaired autophagy
|
homeostasis/metabolism
|
|
| Find Mice |
Using the International Mouse Strain Resource (IMSR)
Mouse lines carrying:
Krastm4Tyj mutation
(9 available);
any
Kras mutation
(76 available)
Tg(Prm-cre)58Og mutation
(4 available)
|
|
|
mortality/aging
|
|
• mutants die between E9.5 and 11.5
|
embryo
|
|
• at E9.5 vasculature is poorly developed; vasculature has a primitive honeycomb-like network lacking branching vitelline vessels, while large vitelline vessels are absent
|
|
|
• at death, embryos exhibit widespread apoptosis
|
|
|
• embryos show developmental arrest
|
|
|
• marked defect in inner labyrinth layer is observed at E9.5
|
|
|
• fetal blood vessels underlying inner labyrinth layer are absent
|
|
|
• at E9.5, yolk sacs are pale and roughened
|
cellular
|
|
• at death, embryos exhibit widespread apoptosis
|
cardiovascular system
|
|
• fetal blood vessels underlying inner labyrinth layer are absent
|
|
|
• at E9.5 vasculature is poorly developed; vasculature has a primitive honeycomb-like network lacking branching vitelline vessels, while large vitelline vessels are absent
|
|
|
| Find Mice |
Using the International Mouse Strain Resource (IMSR)
Mouse lines carrying:
Mettl3tm1.1Jhha mutation
(0 available);
any
Mettl3 mutation
(39 available)
Tg(Prm-cre)58Og mutation
(4 available)
|
|
|
reproductive system
 N N |
• normal male fertility and seminiferous tubules morphology
|
|
|
| Find Mice |
Using the International Mouse Strain Resource (IMSR)
Mouse lines carrying:
Huwe1tm1Alas mutation
(0 available);
any
Huwe1 mutation
(63 available)
Tg(Prm-cre)58Og mutation
(4 available)
|
|
|
reproductive system
 N N |
• male mice are fertile and show normal spermatogenesis with no significant differences in sperm number, sperm motility or testis weights relative to control males
|
|
|
| Find Mice |
Using the International Mouse Strain Resource (IMSR)
Mouse lines carrying:
Inpp5btm2Nbm mutation
(1 available);
any
Inpp5b mutation
(57 available)
Tg(Prm-cre)58Og mutation
(4 available)
|
|
|
reproductive system
|
N |
• mutant males display normal fertility with normal litter sizes
|
|
|
| Find Mice |
Using the International Mouse Strain Resource (IMSR)
Mouse lines carrying:
Fyco1tm1.1Arte mutation
(0 available);
any
Fyco1 mutation
(44 available)
Tg(CAG-RFP/EGFP/Map1lc3b)1Hill mutation
(1 available)
Tg(Prm-cre)58Og mutation
(4 available)
|
|
|
cellular
|
|
• multiphoton laser scanning microscopy showed a 1.3- and 1.6-fold higher GFP intensity in the whole lens and the anterior lens including lens epithelium, respectively, indicating a reduced autophagic flux in mouse lenses
|
homeostasis/metabolism
|
|
| Find Mice |
Using the International Mouse Strain Resource (IMSR)
Mouse lines carrying:
Snx20tm1Lex mutation
(1 available);
any
Snx20 mutation
(12 available)
Tg(Prm-cre)58Og mutation
(4 available)
|
|
|
normal phenotype
|
|
• mice are overall normal and healthy
|
immune system
|
N |
• mice exhibit normal neutrophil morphology and physiology
|






 N
N




 N
N N
N N
N


