Using Ontology Visualization to Coordinate Cross-species Functional Annotation for
Human Disease Genes
 Mary E. Dolan and Judith A. Blake
Mary E. Dolan and Judith A. Blake
Mouse Genome Informatics [MGI], The Jackson Laboratory, 600 Main St., Bar Harbor, ME 04609 USA
Description:
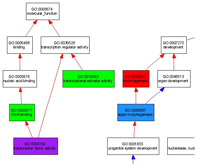
Biomedical ontologies provide a representational system to support the integration and retrieval of biological
knowledge. The Gene Ontology (GO) is widely
used to annotate molecular attributes of genes and provides a common paradigm for comparative functional analysis
research. One way to expand the view of the function of any one gene product is to
compare annotations of genes that share close evolutionary relationships and
are likely to function in similar ways, such as orthologous genes. We are
exploring the power of the GO and OrthoDisease
orthology sets to provide a comprehensive view of annotations coordinated across species by presenting annotations
visualized within the ontology relationship structure. This work describes the
application of our ontology visualization approach to a set of model organism genes
that are orthologous to human disease genes.
View Graphs by OMIM gene or disease id:
Data Sets
 Mary E. Dolan and Judith A. Blake
Mary E. Dolan and Judith A. Blake